Studi perbandingan DNA barcoding untuk Bambu
DOI:
https://doi.org/10.70158/buitenzorg.v1i2.9Abstrak
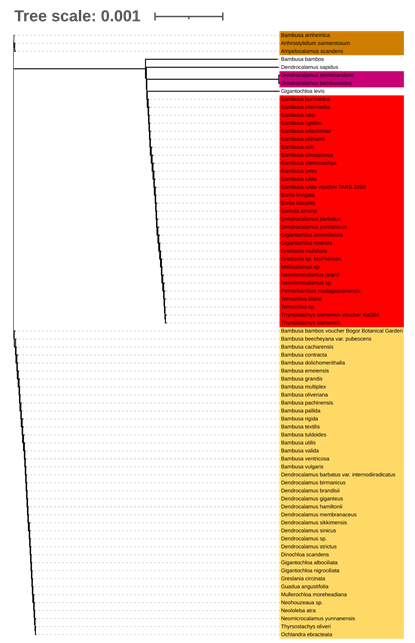
Subfamili Bambusoideae, kelompok penting dalam famili Poaceae, mengandung beragam genus dengan hubungan taksonomi yang kompleks. Penelitian ini bertujuan untuk menjelaskan hubungan filogenetik dan keragaman genetik dalam bambu, dengan fokus pada kegunaan DNA barcoding, yaitu ITS2, matK, dan rbcL, dalam identifikasi spesies bambu. Dengan menganalisis data sekuens dari penanda ini, pohon filogenetik dibangun menggunakan metode kemungkinan maksimum untuk menyimpulkan hubungan evolusi di antara spesies. Hasil penelitian menunjukkan bahwa ITS memberikan resolusi tertinggi untuk identifikasi tingkat spesies, membedakan spesies yang berkerabat dekat secara lebih efektif daripada matK dan rbcL. Sementara matK menunjukkan klasifikasi tingkat genus yang kuat, rbcL dibatasi oleh konservasinya yang tinggi, sehingga lebih cocok untuk pengelompokan taksonomi yang lebih luas. Temuan ini berkontribusi pada pemahaman yang lebih baik tentang taksonomi bambu dan menyoroti pentingnya pemilihan penanda berdasarkan resolusi taksonomi yang diperlukan. Penelitian ini juga menekankan penggunaan pelengkap penanda ini untuk memberikan pandangan yang komprehensif tentang filogenetik bambu.
Kata kunci: taksonomi bambu, DNA barcoding, ITS2, matK, rbcL
Unduhan
Referensi
Benton, A. (2015). Priority Species of Bamboo. In: Liese, W., Köhl, M. (eds) Bamboo. Tropical Forestry, vol 10. Springer, Cham. https://doi.org/10.1007/978-3-319-14133-6_2
Bhat, A. R., Hegde, S., Kammar, S. R., Muthamma, M. B., Mudgal, G., Mohan, T. C., Rajulu, C. (2024). Identification and validation of ITS2-specific universal primers for DNA barcoding in plants. bioRxiv, 2024-01 https://doi.org/10.1101/2024.01.05.574284
Cetiz, M.V., Turumtay, E.A., Burnaz, N.A. et al. (2023). Phylogenetic analysis based on the ITS, matK and rbcL DNA barcodes and comparison of chemical contents of twelve Paeonia taxa in Türkiye. Mol Biol Rep 50, 5195–5208. https://doi.org/10.1007/s11033-023-08435-z
Clark, L.G., Dransfield, S., Triplett, J. and Sánchez-Ken, J.G. (2007). Phylogenetic relationships amongthe one-flowered, determinate genera of Bambuseae (Poaceae: Bambusoideae). Aliso: A Journal of Systematic and Floristic Botany, 23(1), 315–332. https://doi.org/10.5642/aliso.20072301.26
Friar, E., & Kochert, G. (1994). A study of genetic variation and evolution of Phyllostachys (Bambusoideae: Poaceae) using nuclear restriction fragment length polymorphisms. Theoret. Appl. Genetics 89, 265–270. https://doi.org/10.1007/BF00225152
Gogoi, B., Bhau, B.S. (2018). DNA barcoding of the genus Nepenthes (Pitcher plant): a preliminary assessment towards its identification. BMC Plant Biol 18, 153. https://doi.org/10.1186/s12870-018-1375-5
Hodkinson, T., Renvoize, S., Chonghaile, G. et al. (2000). A Comparison of ITS Nuclear rDNA Sequence Data and AFLP Markers for Phylogenetic Studies in Phyllostachys (Bambusoideae, Poaceae). J Plant Res 113, 259–269. https://doi.org/10.1007/PL00013936
Jeevitha, S., & Anandan, R. (2024). Identification and classification of medicinal plant Cassia species through DNA barcode to validate phylogenic analysis of Rbcl, Matk, Ycflb, trnH-psba and ITS marker. Ecology, Environment and Conservation, 30(02), 957–964. https://doi.org/10.53550/eec.2024.v30i02.089
Letsiou, S., Madesis, P., Vasdekis, E. P., Montemurro, C., Grigoriou, M. E., Skavdis, G., Moussis, V., Koutelidakis, A. E., Tzakos, A. G. (2024). DNA Barcoding as a Plant Identification Method. Applied Sciences, 14(4), 1415. https://doi.org/10.3390/app14041415
Lonthor, D. W., Miftahudin, Kayat, Julzarika, A., Subehi, L., Iswandono, E., Dima, A. O. M., Dianto, A., Setiawan, F., & Nugraha, M. F. I. (2023). Diversity of aquatic plants in the Rote Dead Sea area, East Nusa Tenggara, Indonesia, based on rbcL marker. Biodiversitas, 24(2), 810–818. https://doi.org/10.13057/biodiv/d240217
Omonhinmin, C. A., Olomukoro, E. E., Onuselogu, C. C., Popoola, J. O., Oyejide, S. O. rbcL gene dataset on intra-specific genetic variability and phylogenetic relationship of Crassocephalum crepidioides (Benth) S. Moore. (Asteraceae) in Nigeria. Data Brief. 2023 May 26;48: 109266. https://doi.org/10.1016/j.dib.2023.109266
Susanti, F., Adharini, R. I., Sari, D. W. K., & Setyobudi, E. (2023). Genetic Diversity of Gracilaria spp. in the Intertidal Zone on the South Coast of Yogyakarta, Indonesia Based on DNA Barcoding with rbcL Marker. HAYATI Journal of Biosciences, 30(5), 907-917. https://doi.org/10.4308/hjb.30.5.907-917
Su’udi, M., Ulum, F. B., Ardiyansah, M., & Fitri, N. E. (2024). Evaluasi Lokus Potensial matK dan ITS2 Untuk DNA Barcoding Anggrek Bulbophyllum lobbii Lindl. Al-Kauniyah: Jurnal Biologi, 17(2), 406–418. https://doi.org/10.15408/kauniyah.v17i2.33897
Utama, M. N., Etikawati, N., Sugiyarto, & Susilowati, A. (2024). New specific primer matK and rbcL region for DNA barcode pitcher plant Nepenthes spathulata. Biodiversitas, 25(6), 2515–2523. https://doi.org/10.13057/biodiv/d250621
Yang, J. B., Yang, H. Q, Li, D. Z., Wong, K. M, & Yang, Y. M. (2010). Phylogeny of Bambusa and its allies (Poaceae: Bambusoideae) inferred from nuclear GBSSI gene and plastid psbA-trnH, rpl32-trnL and rps16 intron DNA sequences. Taxon, 59(4), 1102–1110. https://doi.org/10.1002/TAX.594010
Yong, W. T. L., Mustafa, A. A., Derise, M. R., & Rodrigues, K. F. (2024). DNA barcoding using chloroplast matK and rbcL regions for the identification of bamboo species in Sabah. Advances in Bamboo Science, 7(December 2023), 100073. https://doi.org/10.1016/j.bamboo.2024.100073
Whitley, B.S., Li, Z., Jones, L., de Vere, N. (2024). Mega-Barcoding Projects: Delivering National DNA Barcoding Initiatives for Plants. In: DeSalle, R. (eds) DNA Barcoding. Methods in Molecular Biology, vol 2744. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3581-0_27
Unduhan
Diterbitkan
Cara Mengutip
Terbitan
Bagian
Lisensi
Hak Cipta (c) 2024 Buitenzorg: Journal of Tropical Science

Artikel ini berlisensiCreative Commons Attribution-ShareAlike 4.0 International License.
The article is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (CC BY-SA), which allows both Authors and Readers to copy and distribute the material in any format or medium, as well as modify and create derivative works from it for any purpose, provided that appropriate credit is given (by citing the article or content), a link to the license is provided, and it is indicated if any changes were made. If the material is modified or used to create derivative works, the contributions must be distributed under the same license as the original.







